
TMIC and the Metabolomics Society
Issue 42 - February 2015
CONTENTS:
Online version of this newsletter:
http://www.metabonews.ca/Feb2015/MetaboNews_Feb2015.htm
 |
| Published
in partnership between TMIC and the Metabolomics Society Issue 42 - February 2015 |
|
CONTENTS: |
|
|
 |
|
 |
|
 |
| Metanomics Health GmbH |
Chenomx
Inc. |
mzCloud
Mass Spectral Database |
 |
Metabolomics Society News |



| |
Statistical Spotlight
|
 |
Quick Overview
1. How to use BioStatFlow

2. BioStatFlow is based on a typical workflow as shown
below:
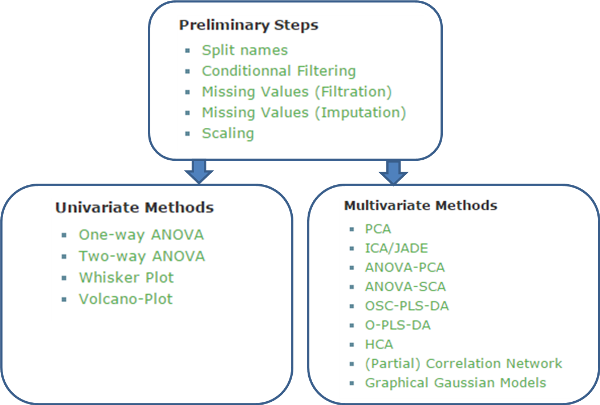
Figure 2. The default workflow implemented in BioStatFlow.
First, to normalize the dataset, a set of analysis is proposed
as a static sequence. At this stage, users must follow the
sequence order. In some cases, because of experimental issues
with the analytical equipment, the levels of some analytical
variables (features) cannot be determined. In other cases where
users want to compare different experiments, missing value
estimation and data scaling are helpful pre-processing steps.
This latter case is the default use case (default workflow).
Users can choose any additional methods depending on the dataset
and the corresponding experimental design (i.e., factors), in
order to: i) visualize the whole data, ii) reveal biomarkers,
iii) analyze interactions between factors, iv) discriminate
groups, etc.
The input to each step takes the output of the previous step.
If a statistical treatment generates a data table (matrix) as an
output, it will be used as the input for the next step.
Otherwise, if the treatment only generates results (texts and
images) but does not change the input array, the latter will be
directly taken as output.
Each statistical treatment step can be written as an R script
(most common) or as a PERL script, embedding binary tools (such
as MATLAB compiled scripts).
3. Examples of univariate and multivariate output results
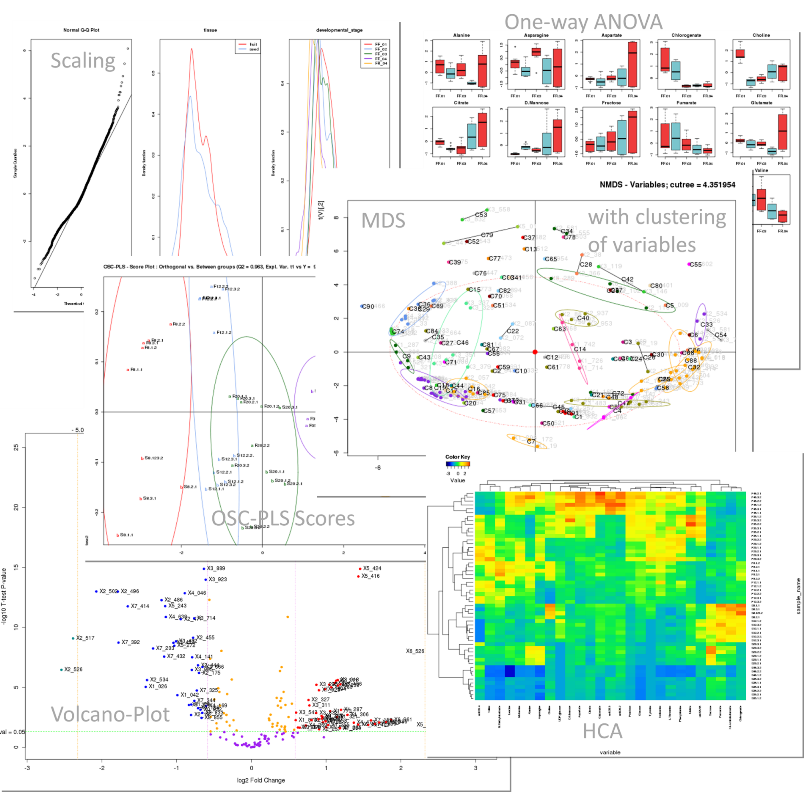
| |
MetaboInterview
|
Head of Metabolomics at the National Institute for Medical Research (NIMR), London, United Kingdom |
|
 |
Biography James MacRae is Head of Metabolomics at the National Institute for Medical Research (NIMR) in London, U.K. (www.nimr.mrc.ac.uk) and will be taking up the same role at The Francis Crick Institute (www.crick.ac.uk) when it opens in late 2015. James has been in the field of mass spectrometry for over 14 years, with his PhD focusing on glycobiology and in trypanosomatid parasites (at the University of Dundee, U.K.), then moving into apicomplexan parasite metabolomics through his post-doctoral research at the University of Melbourne, Australia. His research interests include development of novel and bespoke mass spectrometry-based metabolomics techniques in order to address a variety of metabolism-based research areas, focusing in particular on host-pathogen interactions and cancer metabolism. |
 |
Metabolomics Current Contents
|
This
section of MetaboNews is supported by: |
 |
MetaboNews |
| 22 Jan 2015 |
Federal Government Supports New
Metabolomics Technology Demonstration Centre
The Honourable Michelle Rempel, Minister of State for Western Economic Diversification, today announced an investment of $2,983,800 towards state-of-the-art metabolomics assessment equipment to strengthen western Canadian companies’ ability to commercialize new discoveries. This announcement reinforces Alberta’s position as a global leader in the emerging metabolomics market enabling Genome Alberta, together with the University of Alberta, to establish the Metabolomics Technology Demonstration Centre. The equipment, a 700 MHz Nuclear Magnetic Resonance machine and a Quadrupole-Time of Flight Mass Spectrometer, will assist companies in the metabolomics sector move biomarkers from the research lab to the medical testing facility. Companies will test, validate, and assemble prototype kits using existing research from the University’s Metabolomics Innovation Centre’s biomarker panels, to cost-effectively create more accurate and less invasive medical tests. In addition to an investment from the National Institute of Nanotechnology, Alberta Innovates Health Solutions, Metabolomic Technologies Inc. (MTI), and Genome Canada are also contributors towards this project. Source: Genome Alberta Genomics Blog |
| 2 Feb 2015 |
Metabolomic Discoveries Launches
Personalized Metabolomics Service Kenkodo Through
Crowdfunding Platform Indiegogo
German biotech company Metabolomic Discoveries announced the launch of personalized metabolomics tool Kenkodo. Through a smartphone app and sampling kit, users will soon be able to improve their well-being. Starting today, Kenkodo is available through the crowdfunding platform Indiegogo. This product offers people the possibility to track and understand the influence of factors, such as nutrition, sports and stress on their metabolism. Through lifestyle changes, shifts in the metabolism become measurable and can be optimized accordingly. Kenkodo builds the biochemical fingerprint of every single participant. Dr. Nicolas Schauer, CEO of Metabolomic Discoveries, is very excited about the development of Kenkodo. “Over the last couple of years we have gained a lot of knowledge and experience in metabolomics technology. It tracks more than 1000 small molecules in human samples. Metabolomics has the potential to resolve many questions regarding your wellness and your general constitution. Kenkodo will empower people to understand the effect of their daily lifestyle habits on their body. And finally, to find a lifestyle that keeps them healthy.” Every human has a different, very individual biochemical fingerprint. It combines information about our body’s building blocks like amino acids or lipids and is the ultimate indicator of our level of well-being. “People respond differently to food, beverages and sports. By analyzing the metabolome of a large group of people, we will be able to make links to their lifestyle and well-being and draw meaningful conclusions, for example about food intake.” says Dr. Josephine Worseck, Head of Business Development at Metabolomic Discoveries. Dr. Isam Haddad, Bioinformatics expert at Metabolomic Discoveries, commented about Kenkodo. “We will build a predictive model that relates your lifestyle habits to your body’s response. Our vision is that people will ultimately be able to recognize the metabolic pattern of a great week and repeat it by using the knowledge of Kenkodo.” Source: Metabolomic Discoveries |
Metabolomics Events
|
| 9-12 Feb 2015 |
3rd Annual Practical Applications
of NMR in Industry Conference (PANIC) The PANIC meeting is intended to address topics that occur daily in industrial, government, and academic research laboratories whose primary task entails the application of NMR to a diverse set of analytical problems. Topics include quantitation, molecular structure characterization, trace component and mixture analysis, and product support for a variety of materials that include small molecules, polymers, heterogeneous mixtures, natural products, biosimilars, polysaccharides, and proteins. The conference provides in-depth discussions of the "nuts and bolts" of basic NMR experiments that accent the underlying best-practices developed to address these everyday problems. It also explores the regulatory aspects of the applications of these experiments. Greater insights will be provided into the less frequently explored techniques used in NMR such as quantitation, chemometrics, automation, relaxometry, and at/on line instrumentation. Attendees will have the opportunity to learn about and discuss applications of NMR to polymers, petroleum, food, agriculture, and nutritional supplements that are rarely discussed at other NMR meetings. Ample opportunity will be given to Network with people who are experts in inventing NMR experiments to solve real world problems in a timely and efficient manner that is demanded by working in the venue of product delivery. For more details, visit http://www.panicnmr.com. |
| 16-20 Feb 2015 |
EMBO Practical Course on
Metabolomics Bioinformatics for Life Scientists Organizers
Participation: Open application with
selection |
| 3-6 Mar 2015 |
Introduction to Integrative Omics For more details, visit http://www.ebi.ac.uk/training/course/introduction-integrative-omics. |
| 28 Mar to 1 Apr 2015 |
Plant Metabolism Session at the
ASBMB Annual Meeting |
| 21-22 May 2015 |
2nd Metabolomics - Advances &
Applications in Human Disease Conference Panel: |
| 15-16 Jun 2015 |
Informatics and Statistics for
Metabolomics (2015) A poster announcing this workshop can be found here.The workshop will cover many topics ranging from understanding metabolomics technologies, data collection and analysis, using pathway databases, performing pathway analysis, conducting univariate and multivariate statistics, working with metabolomic databases and exploring chemical databases. Participants will be given various data sets and short assignments to assist with the learning process. Target Audience This course is intended for graduate students, post-doctoral fellows, clinical fellows and investigators who are interested in learning about both bioinformatic and cheminformatic tools to analyze and interpret metabolomics data. Prerequisite: Familiarity with R is required. Familiarity can be gained through online activities. You should be familiar with these R concepts (chapters 1-5) or review the past Statistics tutorials provided by CBW. Apply Now Award Opportunities For further details, visit http://bioinformatics.ca/workshops/2015/informatics-and-statistics-metabolomics-2015. |
| 29 Jun to 2 Jul 2015 |
Metabolomics 2015: 11th Annual
Conference of the International Metabolomics Society You are invited to join us for Metabolomics 2015,
the official annual meeting of the Metabolomics
Society. For further details, visit http://metabolomics2015.org. |
Metabolomics Jobs |
This is a resource for advertising positions in
metabolomics. If you have a job you would like posted in this
newsletter, please email Ian Forsythe (metabolomics.innovation@gmail.com).
Job postings will be carried for a maximum of 4 issues (8
weeks) unless the position is filled prior to that date.
Jobs Offered
| Job Title | Employer | Location | Posted | Closes | Source |
|---|---|---|---|---|---|
| Computational Metabolomics Professor |
Pennsylvania State University |
State College, Pennsylvania, USA | 26-Jan-2015 |
Metabolomics Society Jobs |
|
| Postdoctoral Fellow in Environmental
Metabolomics |
University of Toronto |
Toronto, Canada | 8-Jan-2015 |
28-Feb-2015 |
naturejobs.com |
| Vice Chancellor's Postdoctoral Fellowship |
RMIT University |
Melbourne, Australia | 18-Dec-2014 |
8-Feb-2015 |
RMIT University |
| Vice Chancellor's Research & Senior
Research Fellowships |
RMIT University |
Melbourne, Australia | 18-Dec-2014 |
8-Feb-2015 |
RMIT University |
|
Ian J. Forsythe, M.Sc.
MetaboNewsEditor Department of
Computing Science
University of Alberta 221 Athabasca Hall Edmonton, AB, T6G 2E8, Canada Email: metabolomics.innovation@gmail.com Website: http://www.metabonews.ca LinkedIn: http://ca.linkedin.com/in/iforsythe Twitter: http://twitter.com/MetaboNews Google+: https://plus.google.com/118323357793551595134 Facebook: http://www.facebook.com/metabonews |
This newsletter is published in
partnership between The Metabolomics Innovation Centre
(TMIC, http://www.metabolomicscentre.ca/) and the Metabolomics
Society (http://www.metabolomicssociety.org)
for the benefit of the worldwide metabolomics
community.
|