The
NIH West Coast Metabolomics Center
In September 2013, the NIH Common Funds has announced that three new regional comprehensive metabolomic resource cores will be funded to serve the U.S. academic community with broad services in metabolomics, ranging from flux studies to multi-platform data acquisitions, pathway mapping and interpreting metabolomic data. The first three of these metabolomic service centers were funded 12 months ago at University of Michigan at Ann Arbor, Research Triangle International (Durham, North Carolina) and UC Davis (California). As central repository, a metabolomic workbench and data center has been established at UC San Diego (California) in addition to projects aimed at fostering training in metabolomics, specific metabolomic research projects and two centers for custom synthesis of metabolites, see http://commonfund.nih.gov/metabolomics/.
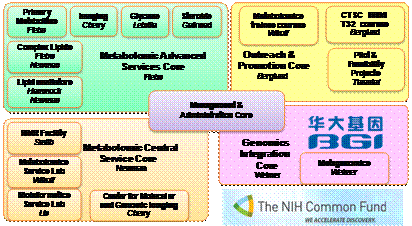 The
West Coast Metabolomics Center (WCMC, see http://metabolomics.ucdavis.edu) may
serve as example how these regional metabolomic service centers function. In a
fast developing field such as metabolomics, services need to constantly evolve
according to new technical developments as well as needs expressed by the
scientific community. Consequently, the WCMC provides capacity for
fee-for-service projects (‘core services’) in addition to direct interaction
and collaboration with metabolomic research laboratories (‘advanced services’ and
‘genomic integration’). Within the core services, there are four major units:
the central service laboratory based on mass spectrometry, the imaging
laboratory, the bioinformatics service unit and the NMR service laboratory. From
August 2012 to June 2013, the central service core alone has provided
metabolomic data for over 12,000 samples in 165 projects. Statistics, pathway
mapping and network analyses are performed on request as hourly services.
The
West Coast Metabolomics Center (WCMC, see http://metabolomics.ucdavis.edu) may
serve as example how these regional metabolomic service centers function. In a
fast developing field such as metabolomics, services need to constantly evolve
according to new technical developments as well as needs expressed by the
scientific community. Consequently, the WCMC provides capacity for
fee-for-service projects (‘core services’) in addition to direct interaction
and collaboration with metabolomic research laboratories (‘advanced services’ and
‘genomic integration’). Within the core services, there are four major units:
the central service laboratory based on mass spectrometry, the imaging
laboratory, the bioinformatics service unit and the NMR service laboratory. From
August 2012 to June 2013, the central service core alone has provided
metabolomic data for over 12,000 samples in 165 projects. Statistics, pathway
mapping and network analyses are performed on request as hourly services.
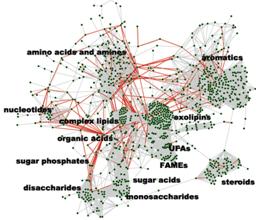 Advancements in metabolomics as well as direct
scientific collaborations are performed in faculty-driven research
laboratories, specifically in glycans (Dr Lebrilla), primary metabolism/complex lipids (Dr Fiehn), oxylipins and lipid mediators (Dr Newman), steroids (Dr Gaikwad), eicosanoids (Dr
Hammock) and genomics (Dr Weimer). Overall, more than 1,000 metabolites can be
targeted in various tissues, cells and biofluids
using these platforms (see network graph). The metabolomic laboratories are
also the primary points of interaction for the yearly U.S.-wide competition for
‘Pilot and Feasibility Grant Awards’ which provide metabolomic services for
NIH-eligible biologists and clinicians for up to $50,000 per project. Seven
projects have been selected in 2013 by the NIH peer review system, of which
only one was submitted by a UC Davis researcher.
Advancements in metabolomics as well as direct
scientific collaborations are performed in faculty-driven research
laboratories, specifically in glycans (Dr Lebrilla), primary metabolism/complex lipids (Dr Fiehn), oxylipins and lipid mediators (Dr Newman), steroids (Dr Gaikwad), eicosanoids (Dr
Hammock) and genomics (Dr Weimer). Overall, more than 1,000 metabolites can be
targeted in various tissues, cells and biofluids
using these platforms (see network graph). The metabolomic laboratories are
also the primary points of interaction for the yearly U.S.-wide competition for
‘Pilot and Feasibility Grant Awards’ which provide metabolomic services for
NIH-eligible biologists and clinicians for up to $50,000 per project. Seven
projects have been selected in 2013 by the NIH peer review system, of which
only one was submitted by a UC Davis researcher.
Outreach to the scientific community is organized together with the UC Davis Clinical Translational Science Center, CTSC. Drs. Tarantal and Berglund direct the pilot grant process in addition to training of clinical researchers in metabolomics within the CTSC courses, workshops and seminars. The West Coast Metabolomics Center has organized an inaugural metabolomics symposium, a microbial metabolism meeting (with hands-on training workshop) and a cancer metabolism conference (in coordination with the UC Davis Comprehensive Cancer Center) with a total of 18 invited speakers. For next year, meetings are being organized for ‘metabolism in animal models’ and computational metabolomics. In addition, 21 national and international guest speakers have discussed their work at the monthly metabolomic seminar series. Intensive training is also provided via courses that are organized by the West Coast Metabolomics Center. This month, the first two-week long course “International Sessions in Metabolomics” has ended with 20 participants from 10 countries who have received theoretical and hands-on instructions in the computer lab, where each course participant actively processed metabolomic data, identified metabolites and performed statistical and pathway analyses of samples that were acquired by themselves in the MS-based metabolomic wet lab.